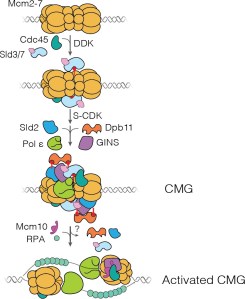
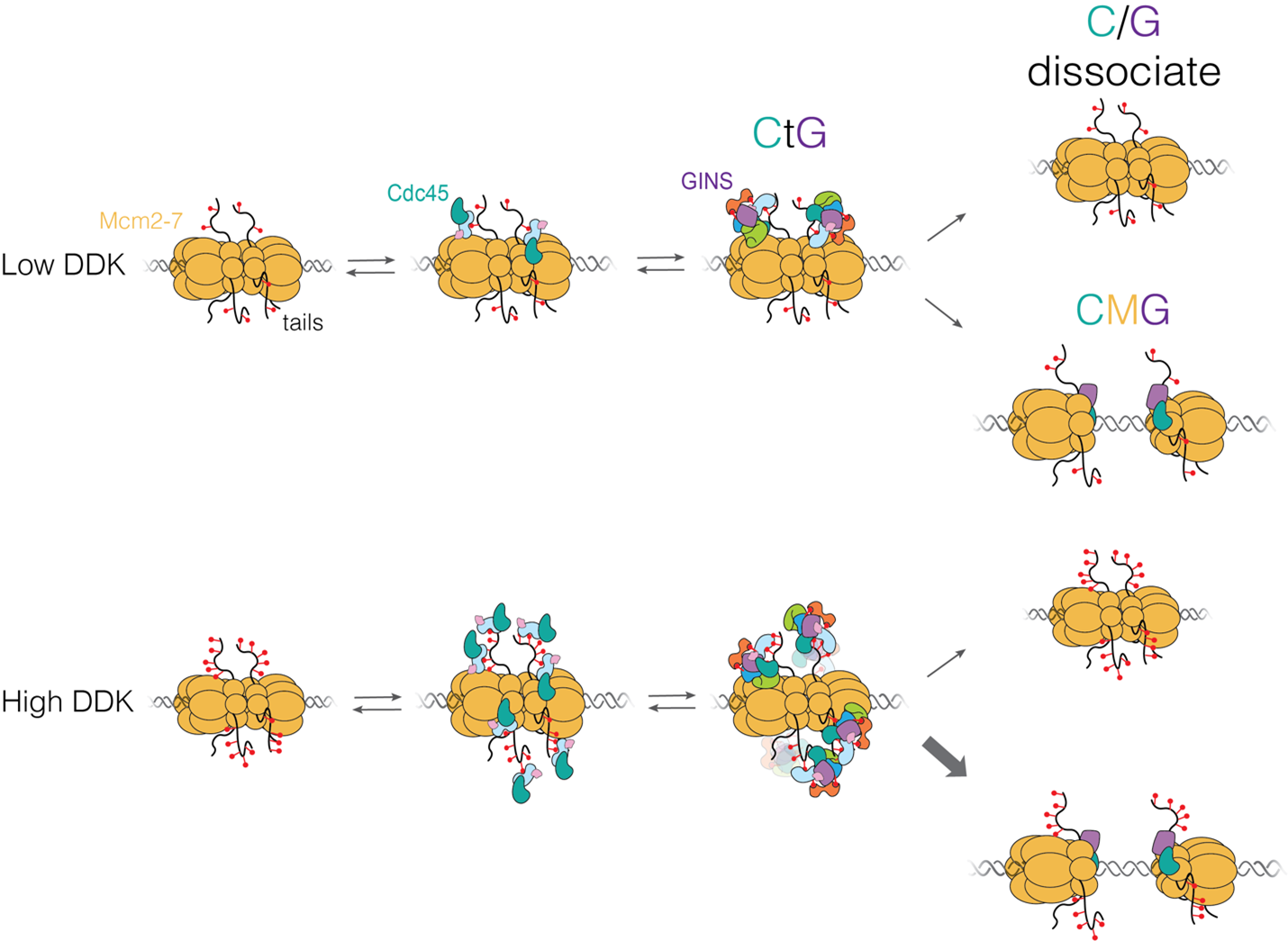
“Each time a eukaryotic cell divides (by mitosis) it must duplicate its chromosomal DNA exactly once to ensure that one full copy is passed to each resulting cell. Both under-replication or over-replication result in genome instability and disease or cell death. A key mechanism to prevent over-replication is the temporal separation of loading of the replicative DNA helicase [Mcm2-7] at origins of replication and activation of these same helicases during the cell division cycle.” Helicase loading is performed by the origin replication complex (ORC), a multi-subunit ATPase. In this study, Audra Amasino and Shalini Gupta from Steve Bell’s lab at MIT, working with Larry Friedman from the Gelles lab at Brandeis, define the mechanism by which cell cycle-dependent phosphorylation of the ORC inhibits helicase loading. Loading is a multi-step process and several steps are inhibited by phosphorylation, presumably helping to ensure that loading is completely suppressed during the S phase of the cell cycle during which the helicases are activated.
Amasino A., et al. Regulation of replication origin licensing by ORC phosphorylation reveals a two-step mechanism for Mcm2-7 ring closing. PNAS, 120, e2221484120 (2023).

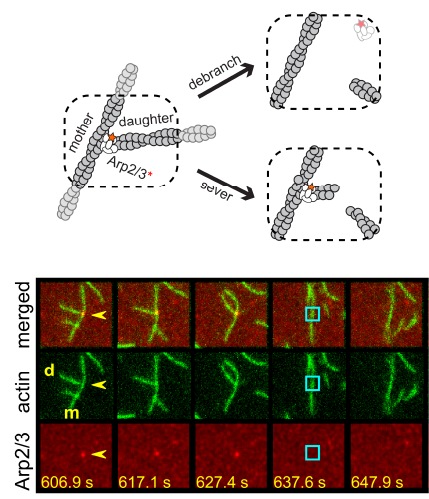 Two separate members of the larger ADF-homology family of proteins, glia maturation factor (GMF) and cofilin, have been implicated in promoting debranching. In this paper, Gelles lab member Johnson Chung, in collaboration with Jeff and with Bruce Goode from the Brandeis Biology Dept., used multi-wavelength single molecule florescence microscopy and quantitative kinetic analysis to define the mechanisms by which these proteins promote debranching. Dr. Chung shows that “cofilin, like GMF, is an authentic debrancher independent of its filament-severing activity and that the debranching activities of the two proteins are additive. While GMF binds directly to the Arp2/3 complex, cofilin selectively accumulates on branch–junction daughter filaments in tropomyosin-decorated networks just prior to debranching events. Quantitative comparison of debranching rates with the known kinetics of cofilin–actin binding suggests that cofilin occupancy of a particular single actin site at the branch junction is sufficient to trigger debranching. In rare cases in which the order of departure could be resolved during GMF- or cofilin-induced debranching, the Arp2/3 complex left the branch junction bound to the pointed end of the daughter filament, suggesting that both GMF and cofilin can work by destabilizing the mother filament–Arp2/3 complex interface. Taken together, these observations suggest that GMF and cofilin promote debranching by distinct yet complementary mechanisms.”
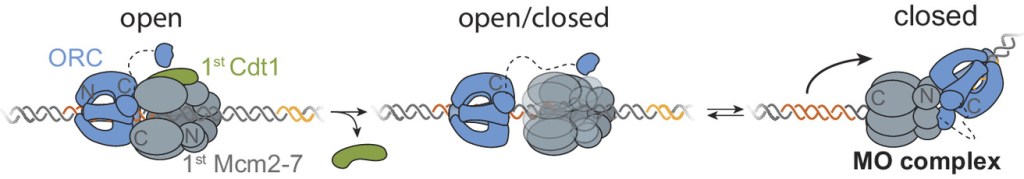
Two separate members of the larger ADF-homology family of proteins, glia maturation factor (GMF) and cofilin, have been implicated in promoting debranching. In this paper, Gelles lab member Johnson Chung, in collaboration with Jeff and with Bruce Goode from the Brandeis Biology Dept., used multi-wavelength single molecule florescence microscopy and quantitative kinetic analysis to define the mechanisms by which these proteins promote debranching. Dr. Chung shows that “cofilin, like GMF, is an authentic debrancher independent of its filament-severing activity and that the debranching activities of the two proteins are additive. While GMF binds directly to the Arp2/3 complex, cofilin selectively accumulates on branch–junction daughter filaments in tropomyosin-decorated networks just prior to debranching events. Quantitative comparison of debranching rates with the known kinetics of cofilin–actin binding suggests that cofilin occupancy of a particular single actin site at the branch junction is sufficient to trigger debranching. In rare cases in which the order of departure could be resolved during GMF- or cofilin-induced debranching, the Arp2/3 complex left the branch junction bound to the pointed end of the daughter filament, suggesting that both GMF and cofilin can work by destabilizing the mother filament–Arp2/3 complex interface. Taken together, these observations suggest that GMF and cofilin promote debranching by distinct yet complementary mechanisms.”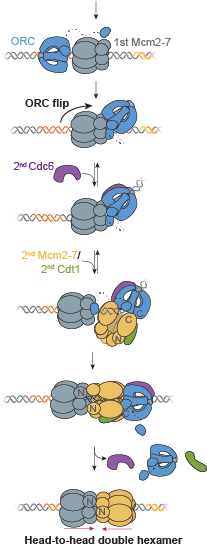 A key event in eukaryotic DNA replication is origin licensing in G1-phase, during which two Mcm2-7 replicative DNA helicases are loaded onto each origin DNA in an inactive, head-to-head fashion. Origin licensing marks every potential origin in a cell, and the opposing orientation of the loaded helicases ensures that they are poised to initiate bidirectional replication when the cell enters S-phase. Although it has long been known that the origin-recognition complex (ORC) binds origin DNA to direct helicase loading, the molecular mechanism by which two oppositely oriented helicases are loaded remains puzzling. Previous biochemical studies found evidence in support of a two-ORC mechanism for helicase loading wherein each of the two Mcm2-7 helicases are recruited by a separate, oppositely oriented ORC molecule. In contrast, single-molecule and cryo-EM approaches observed predominantly one ORC involved in helicase loading, but could not explain how a single ORC could load two oppositely oriented helicases.
A key event in eukaryotic DNA replication is origin licensing in G1-phase, during which two Mcm2-7 replicative DNA helicases are loaded onto each origin DNA in an inactive, head-to-head fashion. Origin licensing marks every potential origin in a cell, and the opposing orientation of the loaded helicases ensures that they are poised to initiate bidirectional replication when the cell enters S-phase. Although it has long been known that the origin-recognition complex (ORC) binds origin DNA to direct helicase loading, the molecular mechanism by which two oppositely oriented helicases are loaded remains puzzling. Previous biochemical studies found evidence in support of a two-ORC mechanism for helicase loading wherein each of the two Mcm2-7 helicases are recruited by a separate, oppositely oriented ORC molecule. In contrast, single-molecule and cryo-EM approaches observed predominantly one ORC involved in helicase loading, but could not explain how a single ORC could load two oppositely oriented helicases.