DNA transcription is the most important nexus of gene regulation in all living organisms. In recent years, single-molecule experiments have given us a new window into the mechanisms of transcription and have revealed novel, previously unsuspected molecular behaviors and mechanisms. Ph.D. student Tim Harden used multi-color single-molecule fluorescence imaging to reveal a completely new transcription cycle for bacterial RNA polymerase. Harden and co-authors showed that an RNA polymerase molecule in vitro frequently (in >90% of transcription events) remains bound to DNA and may again initiate transcription after it has terminated the first round of transcription. Even more unexpectedly, this “secondary initiation” is not restricted to the same RNA. After the first round, the polymerase can scan thousands of basepairs along the DNA and can initiate at a different start site, frequently one that is oriented in the opposite direction and produces an antisense transcript.
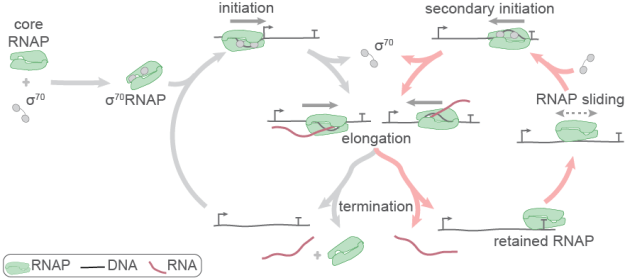
To complement the single-molecule studies in vitro, the manuscript reports new analyses of whole-transcriptome cellular RNAs revealed by the Rend-seq method that measures transcript initiation and termination frequencies across the genome with single basepair resolution. These provide evidence that the new transcription cycle may be responsible for initiating antisense transcription at hundreds of genomic locations in the two widely divergent bacterial species examined. The work defines a new mechanism for the regulated production of antisense RNAs, many of which are now recognized as important agents of gene-specific regulation through control of transcription, mRNA decay, and translation. In addition, the new transcription cycle provides a mechanism through which transcription initiation can be controlled not just through feedback networks involving multiple genes, but also through production of multiple different primary transcripts consequent to a single RNA polymerase-to-DNA recruitment event.
10.1038/s41467-019-14208-9Harden, T.T., et al. Alternative transcription cycle for bacterial RNA polymerase.
Nature Communications 11, 450 (2020).

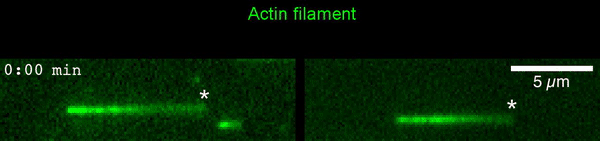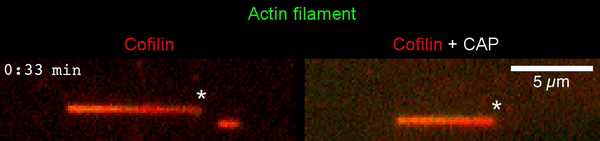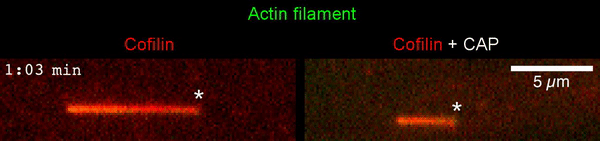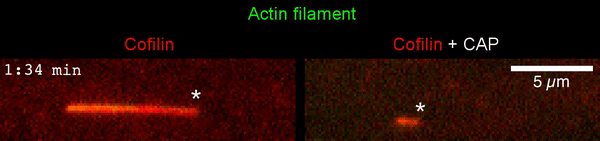 The new study shows that Cofilin and one other protein (Srv2/CAP) intimately collaborate at one end of the actin filament to accelerate subunit dissociation by over 300-fold! These are the fastest rates of actin depolymerization ever observed. Further, these results establish a new paradigm in which a protein that decorates filament sides (Cofilin) works in concert with a protein that binds to filament ends (Srv2/CAP) to produce an activity that is orders of magnitude stronger than the that of either protein alone.”
The new study shows that Cofilin and one other protein (Srv2/CAP) intimately collaborate at one end of the actin filament to accelerate subunit dissociation by over 300-fold! These are the fastest rates of actin depolymerization ever observed. Further, these results establish a new paradigm in which a protein that decorates filament sides (Cofilin) works in concert with a protein that binds to filament ends (Srv2/CAP) to produce an activity that is orders of magnitude stronger than the that of either protein alone.”