The Arp2/3 complex is a multi-component molecular machine that nucleates branched actin filament networks at the leading edge of cells to promote protrusion and at sites of endocytosis to drive membrane invagination. While the process of branched actin nucleation is now well understood (including at mechanistic and structural levels), what is less well understood is how the actin networks are subsequently debranched, or ‘pruned’. Debranching is an absolutely essential step in network remodeling and turnover, which is required for cell motility and endocytosis. The branched actin structures produced by Arp2/3 complex are kinetically stable, with spontaneous dissociation occurring only after tens of minutes to hours, whereas in vivo the branches dissociate in seconds. How is this achieved?
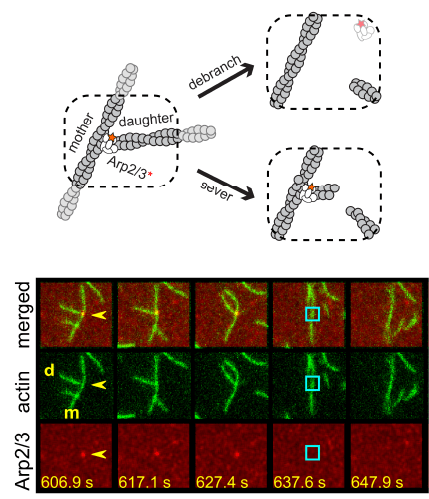 Two separate members of the larger ADF-homology family of proteins, glia maturation factor (GMF) and cofilin, have been implicated in promoting debranching. In this paper, Gelles lab member Johnson Chung, in collaboration with Jeff and with Bruce Goode from the Brandeis Biology Dept., used multi-wavelength single molecule florescence microscopy and quantitative kinetic analysis to define the mechanisms by which these proteins promote debranching. Dr. Chung shows that “cofilin, like GMF, is an authentic debrancher independent of its filament-severing activity and that the debranching activities of the two proteins are additive. While GMF binds directly to the Arp2/3 complex, cofilin selectively accumulates on branch–junction daughter filaments in tropomyosin-decorated networks just prior to debranching events. Quantitative comparison of debranching rates with the known kinetics of cofilin–actin binding suggests that cofilin occupancy of a particular single actin site at the branch junction is sufficient to trigger debranching. In rare cases in which the order of departure could be resolved during GMF- or cofilin-induced debranching, the Arp2/3 complex left the branch junction bound to the pointed end of the daughter filament, suggesting that both GMF and cofilin can work by destabilizing the mother filament–Arp2/3 complex interface. Taken together, these observations suggest that GMF and cofilin promote debranching by distinct yet complementary mechanisms.”
Two separate members of the larger ADF-homology family of proteins, glia maturation factor (GMF) and cofilin, have been implicated in promoting debranching. In this paper, Gelles lab member Johnson Chung, in collaboration with Jeff and with Bruce Goode from the Brandeis Biology Dept., used multi-wavelength single molecule florescence microscopy and quantitative kinetic analysis to define the mechanisms by which these proteins promote debranching. Dr. Chung shows that “cofilin, like GMF, is an authentic debrancher independent of its filament-severing activity and that the debranching activities of the two proteins are additive. While GMF binds directly to the Arp2/3 complex, cofilin selectively accumulates on branch–junction daughter filaments in tropomyosin-decorated networks just prior to debranching events. Quantitative comparison of debranching rates with the known kinetics of cofilin–actin binding suggests that cofilin occupancy of a particular single actin site at the branch junction is sufficient to trigger debranching. In rare cases in which the order of departure could be resolved during GMF- or cofilin-induced debranching, the Arp2/3 complex left the branch junction bound to the pointed end of the daughter filament, suggesting that both GMF and cofilin can work by destabilizing the mother filament–Arp2/3 complex interface. Taken together, these observations suggest that GMF and cofilin promote debranching by distinct yet complementary mechanisms.”
Chung J, et al. Single-molecule analysis of actin filament debranching by cofilin and GMF.
PNAS,119, e2115129119 (2022)

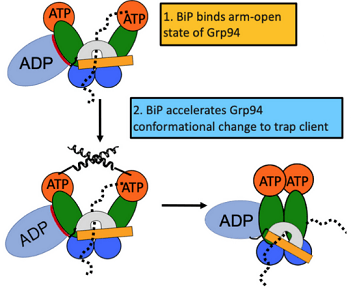 From the article: “The endoplasmic reticulum (ER) is the site at which secreted proteins (such as the hormone insulin) and membrane-bound proteins are folded. ATP-dependent chaperones within the ER help proteins fold. This study describes how two key ER chaperones, BiP and Grp94, work together at a molecular level. BiP binds to Grp94, which enables Grp94 to change conformation and hydrolyze ATP. In short, BiP provides a signal to switch on Grp94 conformational changes that are required to help other proteins fold. This finding helps explain how two chaperones can work together collaboratively in protein folding. Because BiP and Grp94 are members of highly conserved chaperone families, these findings may provide insight into chaperone-assisted protein folding beyond the ER.” This project was a collaboration with members of
From the article: “The endoplasmic reticulum (ER) is the site at which secreted proteins (such as the hormone insulin) and membrane-bound proteins are folded. ATP-dependent chaperones within the ER help proteins fold. This study describes how two key ER chaperones, BiP and Grp94, work together at a molecular level. BiP binds to Grp94, which enables Grp94 to change conformation and hydrolyze ATP. In short, BiP provides a signal to switch on Grp94 conformational changes that are required to help other proteins fold. This finding helps explain how two chaperones can work together collaboratively in protein folding. Because BiP and Grp94 are members of highly conserved chaperone families, these findings may provide insight into chaperone-assisted protein folding beyond the ER.” This project was a collaboration with members of