Transcription initiation is arguably the single most important process for the regulation of gene expression in all organisms. In bacteria, it is a widely accepted dogma that regulated transcription initiation at a promoter sequence controls only the adjacent transcription unit (i.e., a gene or operon). In contrast, this paper demonstrates the feasibility of a new mechanism by which production of multiple RNAs from nearby operons is coupled to the binding of an individual RNA polymerase molecule to a promoter.
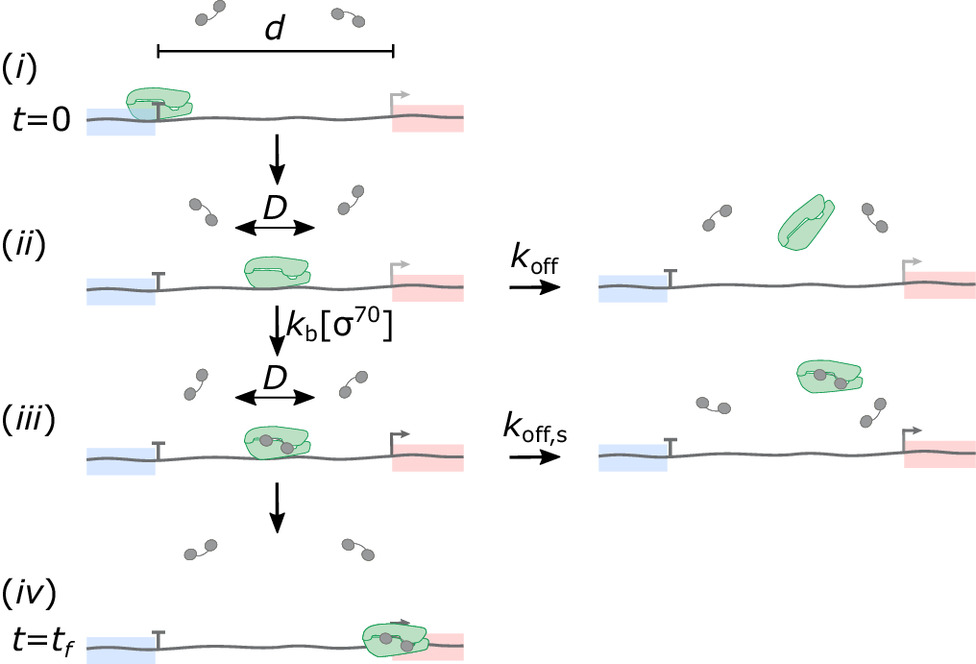
The new mechanism was hypothesized based on the recent discovery in vitro of a post-termination state of the bacterial RNA polymerase in which the polymerase slides randomly on DNA after terminating transcription of one RNA and can reinitiate transcription on a nearby promoter, producing another RNA. Despite these observations, it was unclear whether the hypothesized mechanism could operate efficiently over the time and distance scales necessary to couple nearby operons in a bacterial genome in vivo. Now-graduated Ph.D. student Debora Tenenbaum and collaborators tested this idea by developing a mathematical theory based on a diffusion-to-capture mechanism. The theory quantitatively predicts the efficacy of operon coupling in terms of rate constants that were previously unknown but which the authors measured in single-molecule biophysics experiments. This combination of theory and experiment shows that the mechanism operates on the length and time scales needed to function in bacterial genomes. The results suggest a generalized mechanism that couples the transcription of nearby operons and breaks the paradigm that each binding of RNAP to DNA can produce at most one messenger RNA.
10.1073/pnas.2301402120Tenenbaum D., et al. RNA polymerase sliding on DNA can couple the transcription of nearby bacterial operons. PNAS, 120, e2301402120 (2023)

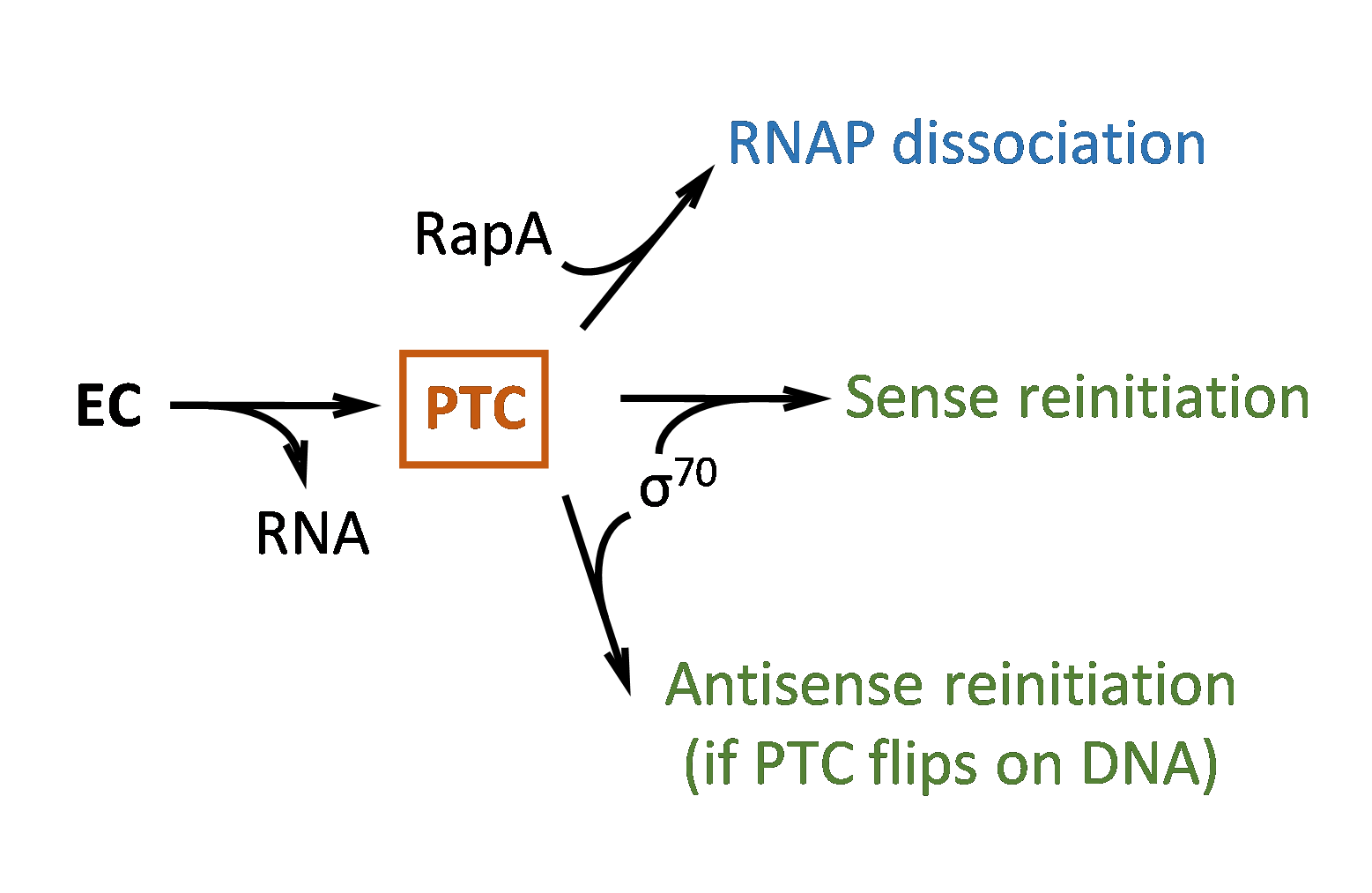 understood in any organism, not even in simple bacteria.
understood in any organism, not even in simple bacteria.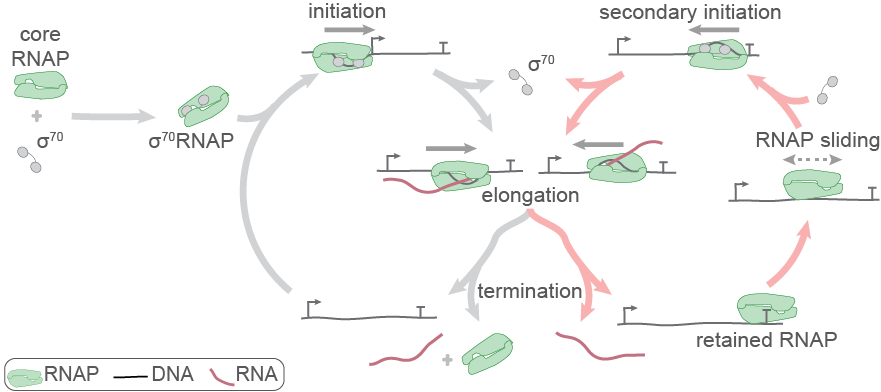
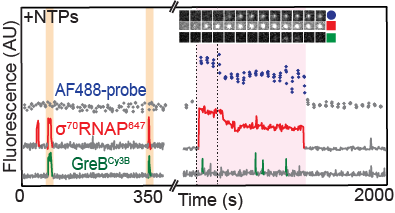 Sarah Stumper and her collaborators used fluorescence correlation spectroscopy and multi-wavelength single-molecule fluorescence colocalization microscopy to show that the SCFs likely repress transcription through an interesting “delayed inhibition” mechanism in which the proteins arrive at DNA already complexed to RNA polymerase and block at a later stage of transcription initiation. The work explains factors that control the relative contributions of the two proteins to regulation and suggests a mechanism by which repression is restricted to ribosomal RNA and other promoters that form short-duration complexes with RNA polymerase.
Sarah Stumper and her collaborators used fluorescence correlation spectroscopy and multi-wavelength single-molecule fluorescence colocalization microscopy to show that the SCFs likely repress transcription through an interesting “delayed inhibition” mechanism in which the proteins arrive at DNA already complexed to RNA polymerase and block at a later stage of transcription initiation. The work explains factors that control the relative contributions of the two proteins to regulation and suggests a mechanism by which repression is restricted to ribosomal RNA and other promoters that form short-duration complexes with RNA polymerase.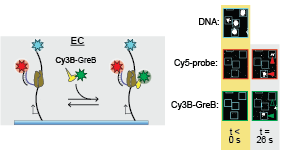 on elongation complex and modulate its activity. In this paper, Larry Tetone, Larry Friedman, and Melissa Osborne, along with their collaborators from the Gelles and
on elongation complex and modulate its activity. In this paper, Larry Tetone, Larry Friedman, and Melissa Osborne, along with their collaborators from the Gelles and