In living cells, messenger RNAs are not manufactured by RNA polymerases (RNAPs) functioning alone. Instead, RNA synthesis is carried out collectively by RNAP together with accessory proteins that associate with the RNAP-containing transcripti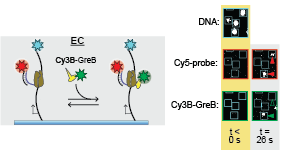 on elongation complex and modulate its activity. In this paper, Larry Tetone, Larry Friedman, and Melissa Osborne, along with their collaborators from the Gelles and Landick labs, used multi-wavelength single-molecule fluorescence methods to for the first time directly observe the dynamic binding and dissociation of an accessory protein with an RNAP during active transcript elongation. The protein, GreB, is important for transcript proofreading in E. coli and other bacteria and is a functional analog of the TFIIS protein in eaukaryotes. “Unexpectedly,” the authors report, “GreB was not selectively recruited to RNAPs requiring its transcript proofreading function. Instead, GreB transiently bound to multiple types of complexes, eventually finding via random search RNAPs that require its activity. The observations suggest a paradigm by which a regulator can act while minimizing obstruction of a binding site that must be shared with other proteins.”
on elongation complex and modulate its activity. In this paper, Larry Tetone, Larry Friedman, and Melissa Osborne, along with their collaborators from the Gelles and Landick labs, used multi-wavelength single-molecule fluorescence methods to for the first time directly observe the dynamic binding and dissociation of an accessory protein with an RNAP during active transcript elongation. The protein, GreB, is important for transcript proofreading in E. coli and other bacteria and is a functional analog of the TFIIS protein in eaukaryotes. “Unexpectedly,” the authors report, “GreB was not selectively recruited to RNAPs requiring its transcript proofreading function. Instead, GreB transiently bound to multiple types of complexes, eventually finding via random search RNAPs that require its activity. The observations suggest a paradigm by which a regulator can act while minimizing obstruction of a binding site that must be shared with other proteins.”
Dynamics of GreB-RNA polymerase interaction allow a proofreading accessory protein to patrol for transcription complexes needing rescue
Larry E. Tetone, Larry J. Friedman, Melisa L. Osborne, Harini Ravi, Scotty Kyzer, Sarah K. Stumper, Rachel A. Mooney, Robert Landick, and Jeff Gelles
PNAS (2017) 114:E1081-E1090.
Resources:
New plasmids reported in this article can be obtained from Addgene