DNA transcription by RNA polymerase II (RNApII) is arguably the process most central to regulation of gene expression in eukaryotic organisms. Regulated transcription requires the formation on DNA of molecular assemblies containing not only RNApII but also dozens of accessory proteins that play pivotal roles in the process. While we know about the structures of some of these assemblies in atomic detail, quantitative understanding of the dynamics and pathways by which the assemblies interconvert and progress through this fundamental gene expression pathway is largely lacking.
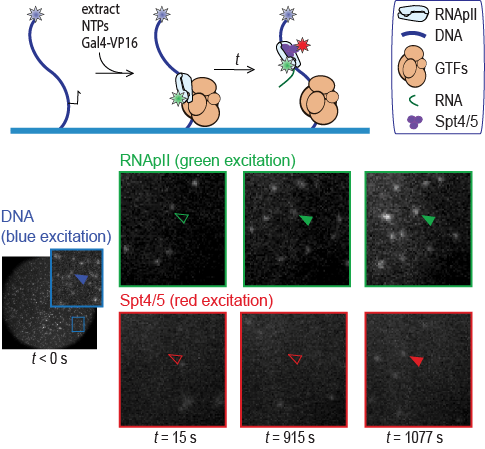
In this study we report single-molecule fluorescence microscopy studies of transcription in yeast nuclear extract, for the first time visualizing and measuring the dynamics of activator-dependent recruitment of RNApII and the central elongation factor Spt4/5 to transcription complexes. Grace Rosen (Jeff Gelles’ labortatory, Brandeis) , Inwha Baek (Steve, Buratowski’s lab, Harvard Medical School), and collaborators elucidated the kinetically significant steps in activated RNApII transcription initiation and show for the first time that Spt4/5 dynamics are tuned to the typical lifetimes of transcription elongation complexes. In addition to these substantive results, our work represents an important methodological advance. As the first application of the CoSMoS (co-localization single-molecule spectroscopy) technique to activated eukaryotic transcription, it demonstrates a general method for elucidating the correlated dynamic interactions of different components of the machinery with initiation and elongation transcription complexes. The approach is likely to find further use in studies of the mechanistic features of RNApII transcription.
https://doi.org/10.1073/pnas.2011224117Rosen, G.A., Baek, I., et al., Dynamics of RNA polymerase II and elongation factor Spt4/5 recruitment during activator-dependent transcription
PNAS

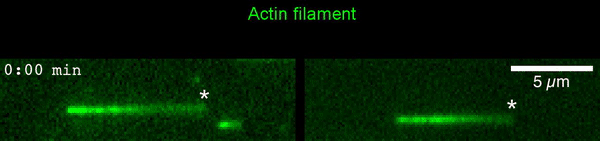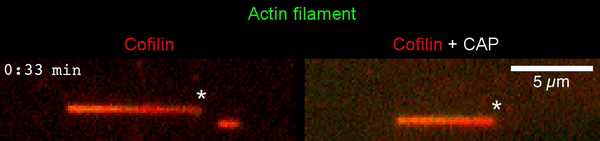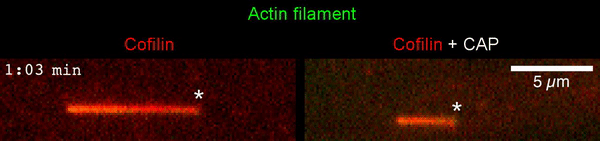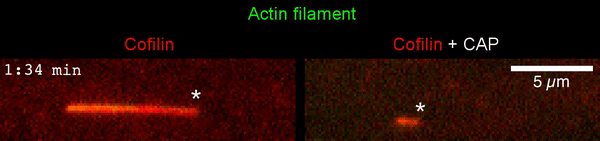 The new study shows that Cofilin and one other protein (Srv2/CAP) intimately collaborate at one end of the actin filament to accelerate subunit dissociation by over 300-fold! These are the fastest rates of actin depolymerization ever observed. Further, these results establish a new paradigm in which a protein that decorates filament sides (Cofilin) works in concert with a protein that binds to filament ends (Srv2/CAP) to produce an activity that is orders of magnitude stronger than the that of either protein alone.”
The new study shows that Cofilin and one other protein (Srv2/CAP) intimately collaborate at one end of the actin filament to accelerate subunit dissociation by over 300-fold! These are the fastest rates of actin depolymerization ever observed. Further, these results establish a new paradigm in which a protein that decorates filament sides (Cofilin) works in concert with a protein that binds to filament ends (Srv2/CAP) to produce an activity that is orders of magnitude stronger than the that of either protein alone.”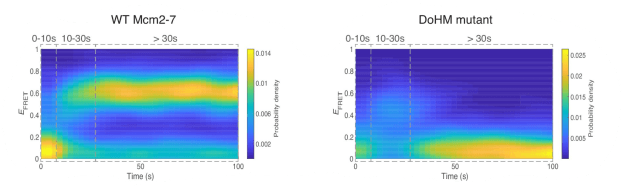
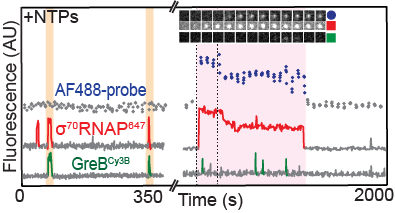 Sarah Stumper and her collaborators used fluorescence correlation spectroscopy and multi-wavelength single-molecule fluorescence colocalization microscopy to show that the SCFs likely repress transcription through an interesting “delayed inhibition” mechanism in which the proteins arrive at DNA already complexed to RNA polymerase and block at a later stage of transcription initiation. The work explains factors that control the relative contributions of the two proteins to regulation and suggests a mechanism by which repression is restricted to ribosomal RNA and other promoters that form short-duration complexes with RNA polymerase.
Sarah Stumper and her collaborators used fluorescence correlation spectroscopy and multi-wavelength single-molecule fluorescence colocalization microscopy to show that the SCFs likely repress transcription through an interesting “delayed inhibition” mechanism in which the proteins arrive at DNA already complexed to RNA polymerase and block at a later stage of transcription initiation. The work explains factors that control the relative contributions of the two proteins to regulation and suggests a mechanism by which repression is restricted to ribosomal RNA and other promoters that form short-duration complexes with RNA polymerase.
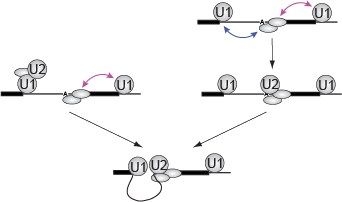
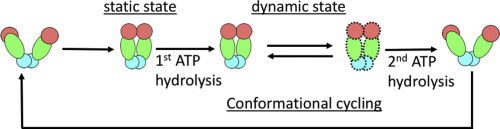
 This work reveals the molecular processes that serve to prevent catastrophic genome damage due to incorrect or mistimed assembly of the replicative machinery.
This work reveals the molecular processes that serve to prevent catastrophic genome damage due to incorrect or mistimed assembly of the replicative machinery.