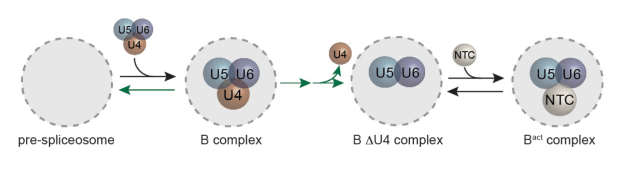
The physical forces that drive oligomerization of soluble proteins are well understood and have been extensively studied. For proteins with transmembrane domains — transport enzymes, for example — oligomerization is often essential for function but its physical basis is less clear. In this project, Janice Robertson devised a new method based on liposome extrusion and single-molecule fluorescence photobleaching analysis to accurately measure the dimer association free energy of a ClC-type chloride ion/hydrogen ion antiporter. (Janice started this work when she was a postdoc in Chris Miller’s lab at Brandeis and later completed the project in her own lab at the University of Iowa.) The study reveals that ClC-ec1 “is one of the strongest membrane protein complexes measured so far, and introduces it as new type of dimerization model to investigate the physical forces that drive membrane protein association in membranes.”
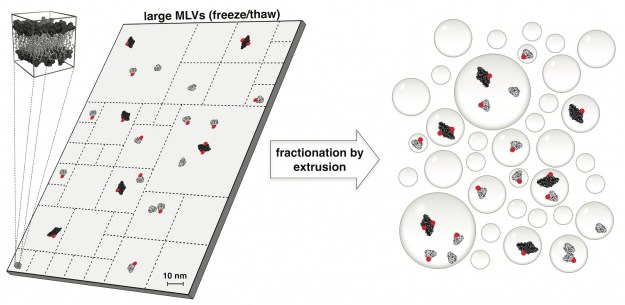
The dimerization equilibrium of a ClC Cl−/H+ antiporter in lipid bilayers
Rahul Chadda, Venkatramanan Krishnamani, Kacey Mersch, Jason Wong, Marley Brimberry, Ankita Chadda, Ludmila Kolmakova-Partensky, Larry J Friedman, Jeff Gelles, and Janice L Robertson
eLife (2016) 5:e17438