“Each time a eukaryotic cell divides (by mitosis) it must duplicate its chromosomal DNA exactly once to ensure that one full copy is passed to each resulting cell. Both under-replication or over-replication result in genome instability and disease or cell death. A key mechanism to prevent over-replication is the temporal separation of loading of the replicative DNA helicase [Mcm2-7] at origins of replication and activation of these same helicases during the cell division cycle.” Helicase loading is performed by the origin replication complex (ORC), a multi-subunit ATPase. In this study, Audra Amasino and Shalini Gupta from Steve Bell’s lab at MIT, working with Larry Friedman from the Gelles lab at Brandeis, define the mechanism by which cell cycle-dependent phosphorylation of the ORC inhibits helicase loading. Loading is a multi-step process and several steps are inhibited by phosphorylation, presumably helping to ensure that loading is completely suppressed during the S phase of the cell cycle during which the helicases are activated.
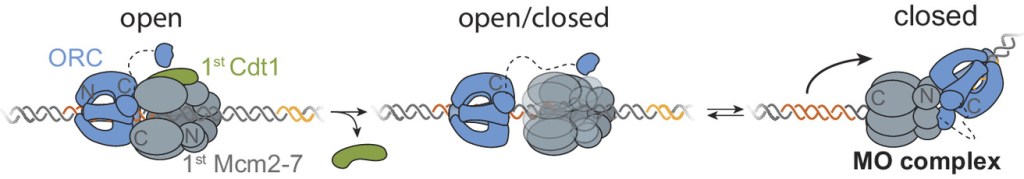
Amasino A., et al. Regulation of replication origin licensing by ORC phosphorylation reveals a two-step mechanism for Mcm2-7 ring closing. PNAS, 120, e2221484120 (2023).

 arina Herlambang, who just defended her dissertation! Karina is moving on with her Ph.D. to work as a scientist at
arina Herlambang, who just defended her dissertation! Karina is moving on with her Ph.D. to work as a scientist at 